Phase 2 Early Access on DARWIN
DARWIN (Delaware Advanced Research Workforce and Innovation Network) is a big data and high performance computing system designed to catalyze Delaware research and education funded by a $1.4 million grant from the National Science Foundation (NSF). This award will establish the DARWIN computing system as an XSEDE Level 2 Service Provider in Delaware contributing 20% of DARWIN's resources to XSEDE: Extreme Science and Engineering Discovery Environment. DARWIN has 105 compute nodes with a total of 6672 cores, 22 GPUs, 100TB of memory, and 1.2PB of disk storage. See architecture descriptions for complete details.
What is Phase 2 Early Access?
Phase 2 early access on DARWIN we will be transitioning to an allocation-based accounting where usage will be tracked against an allocated limit. During this testing period beginning Monday, April 26, 2021, the intent is for research groups currently on DARWIN with experience and established HPC workflows on Caviness to continue testing over the next four weeks unless we encounter problems and need to extend it for further debugging and testing. The following criteria has been defined for Phase 2 early access
- All research groups (PIs only) must submit an application for a startup allocation to be able to submit jobs on DARWIN during Phase 2 early access. Again priority is being given to those participating in the first early access.
- The workgroup unsponsored will no longer be available for job submission and has been changed to read-only. All those granted a startup allocation will be notified with their new workgroup and should move any files from
/lustre/unsponsored/users/<uid>into the new workgroup directory or move them to alternative storage. - All those granted a startup allocation must be willing to provide feedback to IT during the Phase 2 early access period.
We will continue to add accounts during Phase 2 early access only if a startup allocation has been granted and the PI approves the account request.
During DARWIN’s Phase 2 early access period, it is expected that ALL users on the system continue to abide by and follow the University of Delaware’s responsible computing guidelines.
Configuration
Overview
The DARWIN cluster is being set up to be very similar to the existing Caviness cluster, and will be familiar to those currently using Caviness.
An HPC system always has one or more public-facing systems known as login nodes. The login nodes are supplemented by many compute nodes which are connected by a private network. One or more head nodes run programs that manage and facilitate the functioning of the cluster. (In some clusters, the head node functionality is present on the login nodes.) Each compute node typically has several multi-core processors that share memory. Finally, all the nodes share one or more filesystems over a high-speed network.
Login nodes
Login (head) nodes are the gateway into the cluster and are shared by all cluster users. Their computing environment is a full standard variant of Linux configured for scientific applications. This includes command documentation (man pages), scripting tools, compiler suites, debugging/profiling tools, and application software. In addition, the login nodes have several tools to help you move files between the HPC filesystems and your local machine, other clusters, and web-based services.
If your work requires highly interactive graphics and animations, these are best done on your local workstation rather than on the cluster. Use the cluster to generate files containing the graphics information, and download them from the HPC system to your local system for visualization.
When you use SSH to connect to darwin.hpc.udel.edu your computer will choose one of the login (head) nodes at random. The default command line prompt clearly indicates to which login node you have connected: for example, [traine@login00.darwin ~]$ is shown for account traine when connected to login node login00.darwin.hpc.udel.edu.
screen or tmux utility to preserve your session after logout.
Compute nodes
There are many compute nodes with different configurations. Each node consists of multi-core processors (CPUs), memory, and local disk space. Nodes can have different OS versions or OS configurations, but this document assumes all the compute nodes have the same OS and almost the same configuration. Some nodes may have more cores, more memory, GPUs, or more disk.
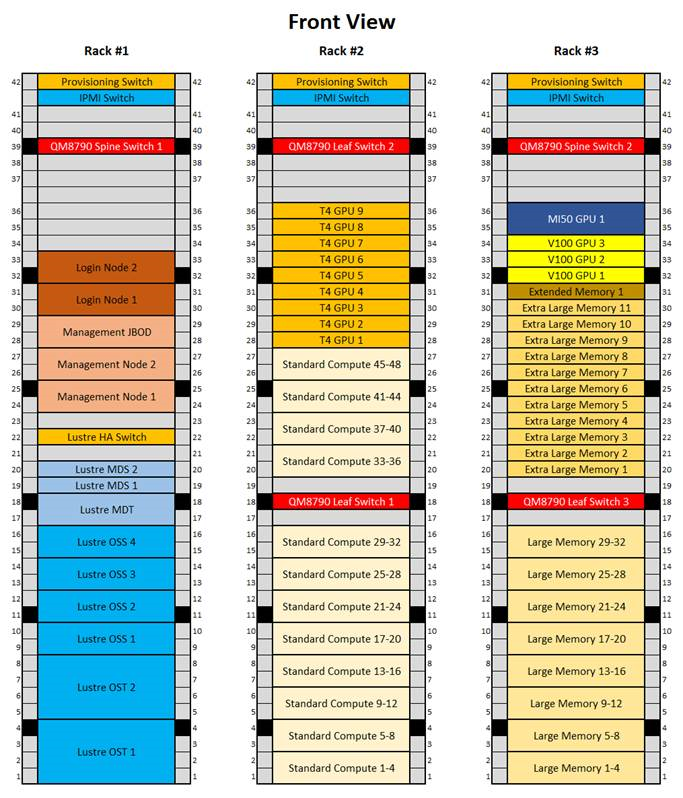
All compute nodes are now available and configured for use. Each compute node has 64 cores, so the compute resources available at initial early access will be the following:
| Compute Node | Number of Nodes | Total Cores | Memory per Node | Total Memory | Total GPUs |
|---|---|---|---|---|---|
| Standard | 48 | 3,072 | 512 GiB | 24 TiB | |
| Large Memory | 32 | 2,048 | 1024 GiB | 32 TiB | |
| Extra-Large Memory | 11 | 704 | 2,048 GiB | 22 TiB | |
| nVidia-T4 | 9 | 576 | 512 GiB | 4.5 TiB | 9 |
| nVidia-V100 | 3 | 144 | 768 GiB | 2.25 TiB | 12 |
| AMD-MI50 | 1 | 64 | 512 GiB | .5 TiB | 1 |
| Extended Memory | 1 | 64 | 1024 GiB + 2.73 TiB1) | 3.73 TiB | |
| Total | 105 | 6,672 | 88.98 TiB | 22 |
The standard Linux on the compute nodes are configured to support just the running of your jobs, particularly parallel jobs. For example, there are no man pages on the compute nodes. All compute nodes will have full development headers and libraries.
Commercial applications, and normally your programs, will use a layer of abstraction called a programming model. Consult the cluster specific documentation for advanced techniques to take advantage of the low level architecture.
Storage
Home filesystem
Each DARWIN user receives a home directory (/home/<uid>) that will remain the same during and after the early access period. This storage has slower access with a limit of 20 GiB. It should be used for personal software installs and shell configuration files.
Lustre high-performance filesystem
Lustre is designed to use parallel I/O techniques to reduce file-access time. The Lustre filesystems in use at UD are composed of many physical disks using RAID technologies to give resilience, data integrity, and parallelism at multiple levels. There is approximately 1.1 PiB of Lustre storage available on DARWIN. It uses high-bandwidth interconnects such as Mellanox HDR100. Lustre should be used for storing input files, supporting data files, work files, and output files associated with computational tasks run on the cluster.
During Phase 2 early access period:
- Each allocation will be assigned a workgroup storage in the Lustre directory (
/lustre/«workgroup»/). - Each workgroup storage will have a users directory (
/lustre/«workgroup»/users/«uid») for each user of the workgroup to be used as a personal directory for running jobs and storing larger amounts of data. - Each workgroup storage will have a software and VALET directory (
/lustre/«workgroup»/sw/and/lustre/«workgroup»/sw/valet) all allow users of the workgroup to install software and create VALET package files that need to be shared by others in the workgroup. - There will be a 1 TiB quota limit for the workgroup storage.
Local filesystems
Each node has an internal, locally connected disk. Its capacity is measured in terabytes. Each compute node on DARWIN has a 1.75 TiB SSD local scratch filesystem disk. Part of the local disk is used for system tasks such memory management, which might include cache memory and virtual memory. This remainder of the disk is ideal for applications that need a moderate amount of scratch storage for the duration of a job's run. That portion is referred to as the node scratch filesystem.
Each node scratch filesystem disk is only accessible by the node in which it is physically installed. The job scheduling system creates a temporary directory associated with each running job on this filesystem. When your job terminates, the job scheduler automatically erases that directory and its contents.
More detailed information about DARWIN storage and quotas can be found on the sidebar under Storage.
Software
A list of installed software that IT builds and maintains for DARWIN users can be found by logging into DARWIN and using the VALET command vpkg_list.
Documentation for all software is organized in alphabetical order on the sidebar under Software. There will likely not be a details by cluster for DARWIN, however referring to Caviness should still be applicable for now.
There will not be a full set of software during early access and testing, but we will be continually installing and updating software. Installation priority will go to compilers, system libraries, and highly utilized software packages. Please DO let us know if there are packages that you would like to use on DARWIN, as that will help us prioritize user needs, but understand that we may not be able to install software requests in a timely manner.
Please review the following documents if you are planning to compile and install your own software
- High Performance Computing (HPC) Tuning Guide for AMD EPYC™ 7002 Series Processors guide for getting started tuning AMD 2nd Gen EPYC™ Processor based systems for HPC workloads. This is not an all-inclusive guide and some items may have similar, but different, names in specific OEM systems (e.g. OEM-specific BIOS settings). Every HPC workload varies in its performance characteristics. While this guide is a good starting point, you are encouraged to perform your own performance testing for additional tuning. This guide also provides suggestions on which items should be the focus of additional, application-specific tuning (November 2020).
- HPC Tuning Guide for AMD EPYC™ Processors guide intended for vendors, system integrators, resellers, system managers and developers who are interested in EPYC system configuration details. There is also a discussion on the AMD EPYC software development environment, and we include four appendices on how to install and run the HPL, HPCG, DGEMM, and STREAM benchmarks. The results produced are ‘good’ but are not necessarily exhaustively tested across a variety of compilers with their optimization flags (December 2018).
- AMD EPYC™ 7xx2-series Processors Compiler Options Quick Reference Guide, however we do not have the AOCC compiler (with Flang - Fortran Front-End) installed on DARWIN.
Scheduler
DARWIN will being using the Slurm scheduler like Caviness, and is the most common scheduler among XSEDE resources. Slurm on DARWIN is configured as fairshare with each user being giving equal shares to access the current HPC resources available on DARWIN.
Queues (Partitions)
During Phase 2 early access partitions have been created to align with allocation requests moving forward based on different node types. There will be no default partition, and only specify one partition at a time. It is not possible to specify multiple partitions using Slurm to span different node types.
See Queues on the sidebar for detailed information about the available partitions on DARWIN.
We fully expect these limits to be changed and adjusted during the early access period.
Run Jobs
In order to schedule any job (interactively or batch) on the DARWIN cluster, you must set your workgroup to define your cluster group. For Phase 2 early access, each research group has been assigned a unique workgroup. Each research group should have received this information in a welcome email for Phase 2 early access. For example,
workgroup -g it_css
will enter the workgroup for it_css. You will know if you are in your workgroup based on the change in your bash prompt. See the following example for user traine
[traine@login00.darwin ~]$ workgroup -g it_css [(unsponsored:traine)@login00.darwin ~]$ printenv USER HOME WORKDIR WORKGROUP WORKDIR_USER traine /home/1201 /lustre/it_css it_css /lustre/it_css/users/1201 [(it_css:traine)@login00.darwin ~]$
Now we can use salloc or sbatch as long as a partition is specified as well to submit an interactive or batch job respectively. See DARWIN Run Jobs, Schedule Jobs and Managing Jobs wiki pages for more help about Slurm including how to specify resources and check on the status of your jobs.
/opt/shared/templates/slurm/ for updated or new templates to use as job scripts to run generic or specific applications, designed to provide the best performance on DARWIN.
See Run jobs on the sidebar for detailed information about the running jobs on DARWIN and specifically Schedule job options for memory, time, gpus, etc.
Help
System or account problems
It is important to understand that DARWIN is in a pre-production state. Portions of the cluster are still under construction and may require outages or reboots during this period. IT staff will attempt to provide 24 hours notice to darwin-users Google group for any expected outages, but early access users should recognize that emergency and unexpected outages may occur.
During the early access to DARWIN, IT staff will be focusing on bringing DARWIN into full production mode and will not be able to provide our standard level of support during this time period. It is fully expected that users will report any problems that they may encounter in early access, as your use of this system is in part to help us fully test this new architecture, but we may not be able to resolve all issues until we are in full production mode.
To report a problem or feedback, please submit a Research Computing High Performance Computing (HPC) Clusters Help Request and complete the form including DARWIN and your problem or feedback details in the description field.
Ask or tell the HPC community
hpc-ask is a Google group established to stimulate interactions within UD’s broader HPC community and is based on members helping members. This is a great venue to post a question about HPC, start a discussion, or share an upcoming event with the community. Anyone may request membership. Messages are sent as a daily summary to all group members. This list is archived, public, and searchable by anyone.
Publication and Grant Writing Resources
Please refer to the NSF award information for a proposal or publication to acknowledge use of or describe DARWINS resources.